Chapter 3: More on @transform-ing data¶
See also
Review¶
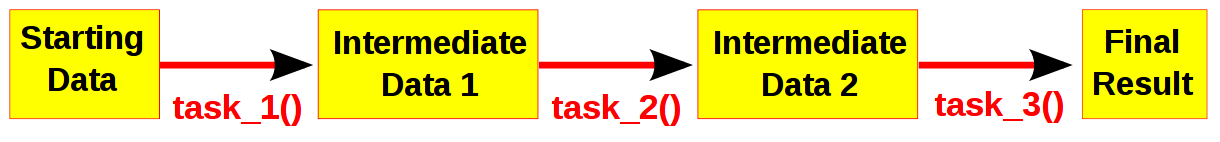
Computational pipelines transform your data in stages until the final result is produced. Ruffus automates the plumbing in your pipeline. You supply the python functions which perform the data transformation, and tell Ruffus how these pipeline stages or task functions are connected together.
Note
The best way to design a pipeline is to:
- write down the file names of the data as it flows across your pipeline
- write down the names of functions which transforms the data at each stage of the pipeline.
Chapter 1: An introduction to basic Ruffus syntax described the bare bones of a simple Ruffus pipeline.
Using the Ruffus @transform decorator, we were able to specify the data files moving through our pipeline so that our specified task functions could be invoked.
This may seem like a lot of effort and complication for something so simple: a couple of simple python function calls we could have invoked ourselves. However, By letting Ruffus manage your pipeline parameters, you will get the following features for free:
- Only out-of-date parts of the pipeline will be re-run
- Multiple jobs can be run in parallel (on different processors if possible)
- Pipeline stages can be chained together automatically. This means you can apply your pipeline just as easily to 1000 files as to 3.
Running pipelines in parallel¶
Even though three sets of files have been specified for our initial pipeline, and they can be processed completely independently, by default Ruffus runs each of them serially in succession.
To ask Ruffus to run them in parallel, all you have to do is to add a multiprocess parameter to pipeline_run:
>>> pipeline_run(multiprocess = 5)In this case, we are telling Ruffus to run a maximum of 5 jobs at the same time. Since we only have three sets of data, that is as much parallelism as we are going to get...
Up-to-date jobs are not re-run unnecessarily¶
A job will be run only if the output file timestamps are out of date. If you ran our example code a second time, nothing would happen because all the work is already complete.
We can check the details by asking Ruffus for more verbose output
>>> pipeline_run(verbose = 4) Task = map_dna_sequence All jobs up to date Task = compress_sam_file All jobs up to date Task = summarise_bam_file All jobs up to date
- Nothing happens because:
- a.sam was created later than a.1.fastq and a.2.fastq, and
- a.bam was created later than a.sam and
- a.statistics was created later than a.bam.
and so on...
- Let us see what happens if we recreated the file a.1.fastq so that it appears as if 1 out of the original data files is out of date
The up to date jobs are cleverly ignored and only the out of date files are reprocessed.
>>> open("a.1.fastq", "w") >>> pipeline_run(verbose=2) Job = [[b.1.fastq, b.2.fastq] -> b.sam] # unnecessary: already up to date Job = [[c.1.fastq, c.2.fastq] -> c.sam] # unnecessary: already up to date Job = [[a.1.fastq, a.2.fastq] -> a.sam] completed Completed Task = map_dna_sequence Job = [b.sam -> b.bam] # unnecessary: already up to date Job = [c.sam -> c.bam] # unnecessary: already up to date Job = [a.sam -> a.bam] completed Completed Task = compress_sam_file Job = [b.bam -> b.statistics, use_linear_model] # unnecessary: already up to date Job = [c.bam -> c.statistics, use_linear_model] # unnecessary: already up to date Job = [a.bam -> a.statistics, use_linear_model] completed Completed Task = summarise_bam_file
Defining pipeline tasks out of order¶
The examples so far assumes that all your pipelined tasks are defined in order. (first_task before second_task). This is usually the most sensible way to arrange your code.
If you wish to refer to tasks which are not yet defined, you can do so by quoting the function name as a string and wrapping it with the indicator class output_from(...) so that Ruffus knowns this is a task name, not a file name
#--------------------------------------------------------------- # # second task # # task name string wrapped in output_from(...) @transform(output_from("first_task"), suffix(".output.1"), ".output2") def second_task(input_files, output_file): with open(output_file, "w"): pass #--------------------------------------------------------------- # # first task # @transform(first_task_params, suffix(".start"), [".output.1", ".output.extra.1"], "some_extra.string.for_example", 14) def first_task(input_files, output_file_pair, extra_parameter_str, extra_parameter_num): for output_file in output_file_pair: with open(output_file, "w"): pass #--------------------------------------------------------------- # # Run # pipeline_run([second_task])You can also refer to tasks (functions) in other modules, in which case the full qualified name must be used:
@transform(output_from("other_module.first_task"), suffix(".output.1"), ".output2") def second_task(input_files, output_file): pass
Multiple dependencies¶
Each task can depend on more than one antecedent simply by chaining to a list in @transform
# # third_task depends on both first_task() and second_task() # @transform([first_task, second_task], suffix(".output.1"), ".output2") def third_task(input_files, output_file): with open(output_file, "w"): passthird_task() depends on and follows both first_task() and second_task(). However, these latter two tasks are independent of each other and can and will run in parallel. This can be clearly shown for our example if we added a little randomness to the run time of each job:
time.sleep(random.random())The execution of first_task() and second_task() jobs will be interleaved and they finish in no particular order:
>>> pipeline_run([third_task], multiprocess = 6) Job = [[job3.a.start, job3.b.start] -> [job3.a.output.1, job3.a.output.extra.1], some_extra.string.for_example, 14] completed Job = [[job6.a.start, job6.b.start] -> [job6.a.output.1, job6.a.output.extra.1], some_extra.string.for_example, 14] completed Job = [[job1.a.start, job1.b.start] -> [job1.a.output.1, job1.a.output.extra.1], some_extra.string.for_example, 14] completed Job = [[job4.a.start, job4.b.start] -> [job4.a.output.1, job4.a.output.extra.1], some_extra.string.for_example, 14] completed Job = [[job5.a.start, job5.b.start] -> [job5.a.output.1, job5.a.output.extra.1], some_extra.string.for_example, 14] completed Completed Task = second_task Job = [[job2.a.start, job2.b.start] -> [job2.a.output.1, job2.a.output.extra.1], some_extra.string.for_example, 14] completedNote
See the example code
@follows¶
If there is some extrinsic reason one non-dependent task has to precede the other, then this can be specified explicitly using @follows:
# # @follows specifies a preceding task # @follows("first_task") @transform(second_task_params, suffix(".start"), [".output.1", ".output.extra.1"], "some_extra.string.for_example", 14) def second_task(input_files, output_file_pair, extra_parameter_str, extra_parameter_num):@follows specifies either a preceding task (e.g. first_task), or if it has not yet been defined, the name (as a string) of a task function (e.g. "first_task").
With the addition of @follows, all the jobs of second_task() start after those from first_task() have finished:
>>> pipeline_run([third_task], multiprocess = 6) Job = [[job2.a.start, job2.b.start] -> [job2.a.output.1, job2.a.output.extra.1], some_extra.string.for_example, 14] completed Job = [[job3.a.start, job3.b.start] -> [job3.a.output.1, job3.a.output.extra.1], some_extra.string.for_example, 14] completed Job = [[job1.a.start, job1.b.start] -> [job1.a.output.1, job1.a.output.extra.1], some_extra.string.for_example, 14] completed Completed Task = first_task Job = [[job4.a.start, job4.b.start] -> [job4.a.output.1, job4.a.output.extra.1], some_extra.string.for_example, 14] completed Job = [[job6.a.start, job6.b.start] -> [job6.a.output.1, job6.a.output.extra.1], some_extra.string.for_example, 14] completed Job = [[job5.a.start, job5.b.start] -> [job5.a.output.1, job5.a.output.extra.1], some_extra.string.for_example, 14] completed Completed Task = second_task
Making directories automatically with @follows and mkdir¶
@follows is also useful for making sure one or more destination directories exist before a task is run.
Ruffus provides special syntax to support this, using the special mkdir indicator class. For example:
# # @follows specifies both a preceding task and a directory name # @follows("first_task", mkdir("output/results/here")) @transform(second_task_params, suffix(".start"), [".output.1", ".output.extra.1"], "some_extra.string.for_example", 14) def second_task(input_files, output_file_pair, extra_parameter_str, extra_parameter_num):Before second_task() is run, the output/results/here directory will be created if necessary.
Globs in the Input parameter¶
- As a syntactic convenience, Ruffus also allows you to specify a glob pattern (e.g. *.txt) in the Input parameter.
- glob patterns will be automatically specify all matching file names as the Input.
- Any strings within Input which contain the letters: *?[] will be treated as a glob pattern.
The first function in our initial Ruffus pipeline example could have been written as:
# # STAGE 1 fasta->sam # @transform("*.fasta", # Input = glob suffix(".fasta"), # suffix = .fasta ".sam") # Output suffix = .sam def map_dna_sequence(input_file, output_file): ""
Mixing Tasks and Globs in the Input parameter¶
glob patterns, references to tasks and file names strings can be mixed freely in (nested) python lists and tuples in the Input parameter.
For example, a task function can chain to the Output from multiple upstream tasks:
@transform([task1, task2, # Input = multiple tasks "aa*.fasta", + all files matching glob "zz.fasta"] + file name suffix(".fasta"), # suffix = .fasta ".sam") # Output suffix = .sam def map_dna_sequence(input_file, output_file): ""In all cases, Ruffus tries to do the right thing, and to make the simple or obvious case require the simplest, least onerous syntax.
If sometimes Ruffus does not behave the way you expect, please write to the authors: it may be a bug!
Chapter 5: Understanding how your pipeline works with pipeline_printout(...) and Chapter 6: Running Ruffus from the command line with ruffus.cmdline will show you how to to make sure that your intentions are reflected in Ruffus code.