Chapter 12: Splitting up large tasks / files with @split¶
See also
Overview¶
A common requirement in computational pipelines is to split up a large task into small jobs which can be run on different processors, (or sent to a computational cluster). Very often, the number of jobs depends dynamically on the size of the task, and cannot be known beforehand.
Ruffus uses the @split decorator to indicate that the task function will produce an indeterminate number of independent Outputs from a single Input.
Example: Calculate variance for a large list of numbers in parallel¶
Suppose we wanted to calculate the variance for 100,000 numbers, how can we parallelise the calculation so that we can get an answer as speedily as possible?
We need to
- break down the problem into manageable chunks
- solve these in parallel, possibly on a computational cluster and then
- merge the partial solutions back together for a final result.
To complicate things, we usually do not want to hard-code the number of parallel chunks beforehand. The degree of parallelism is often only apparent as we process our data.
Ruffus was designed to solve such problems which are common, for example, in bioinformatics and genomics.
A flowchart for our variance problem might look like this:
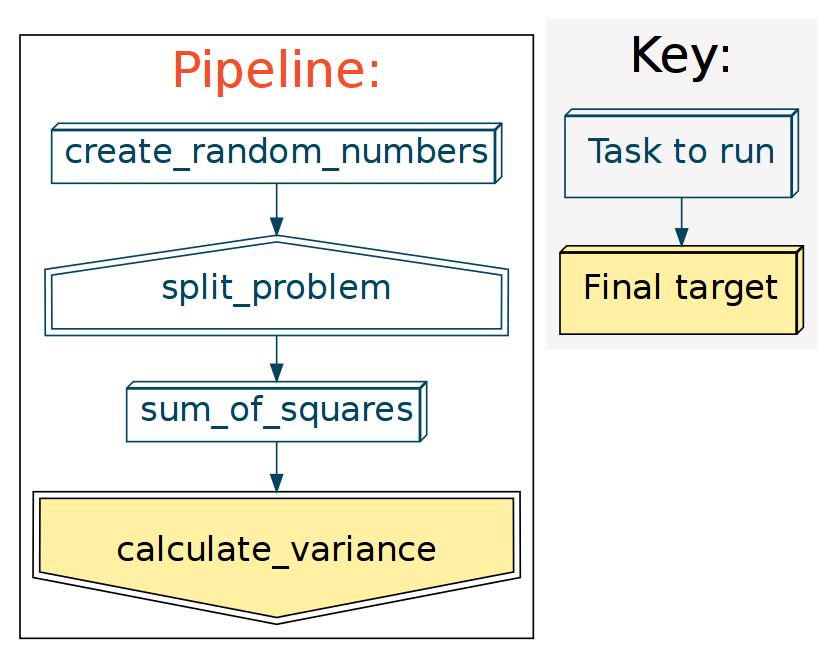
(In this toy example, we create our own starting data in create_random_numbers().)
Output files for @split¶
The Ruffus decorator @split is designed specifically with this run-time flexibility in mind:
@split(create_random_numbers, "*.chunks") def split_problem (input_file_names, output_files): passThis will split the incoming input_file_names into NNN number of outputs where NNN is not predetermined:
The output (second) parameter of @split often contains a glob pattern like the *.chunks above.
Only after the task function has completed, will Ruffus match the Output parameter (*.chunks) against the files which have been created by split_problem() (e.g. 1.chunks, 2.chunks, 3.chunks)
Be careful in specifying Output globs¶
Note that it is your responsibility to keep the Output specification tight enough so that Ruffus does not pick up extraneous files.
You can specify multiple glob patterns to match all the files which are the result of the splitting task function. These can even cover different directories, or groups of file names. This is a more extreme example:
@split("input.file", ['a*.bits', 'b*.pieces', 'somewhere_else/c*.stuff']) def split_function (input_filename, output_files): "Code to split up 'input.file'"
Clean up previous pipeline runs¶
Problem arise when the current directory contains results of previous pipeline runs.
- For example, if the previous analysis involved a large data set, there might be 3 chunks: 1.chunks, 2.chunks, 3.chunks.
- In the current analysis, there might be a smaller data set which divides into only 2 chunks, 1.chunks and 2.chunks.
- Unfortunately, 3.chunks from the previous run is still hanging around and will be included erroneously by the glob *.chunks.
Warning
Your first duty in @split tasks functions should be to clean up
To help you clean up thoroughly, Ruffus initialises the output parameter to all files which match specification.
The first order of business is thus invariably to cleanup ( delete with os.unlink) all files in Output.
#--------------------------------------------------------------- # # split initial file # @split(create_random_numbers, "*.chunks") def split_problem (input_file_names, output_files): """ splits random numbers file into xxx files of chunk_size each """ # # clean up any files from previous runs # #for ff in glob.glob("*.chunks"): for ff in input_file_names: os.unlink(ff)(The first time you run the example code, *.chunks will initialise output_files to an empty list. )
1 to many¶
@split is a one to many operator because its outputs are a list of independent items.
If @split generates 5 files, then this will lead to 5 jobs downstream.
This means we can just connect our old friend @transform to our pipeline and the results of @split will be analysed in parallel. This code should look familiar:
#--------------------------------------------------------------- # # Calculate sum and sum of squares for each chunk file # @transform(split_problem, suffix(".chunks"), ".sums") def sum_of_squares (input_file_name, output_file_name): passWhich results in output like this:
>>> pipeline_run() Job = [[random_numbers.list] -> *.chunks] completed Completed Task = split_problem Job = [1.chunks -> 1.sums] completed Job = [10.chunks -> 10.sums] completed Job = [2.chunks -> 2.sums] completed Job = [3.chunks -> 3.sums] completed Job = [4.chunks -> 4.sums] completed Job = [5.chunks -> 5.sums] completed Job = [6.chunks -> 6.sums] completed Job = [7.chunks -> 7.sums] completed Job = [8.chunks -> 8.sums] completed Job = [9.chunks -> 9.sums] completed Completed Task = sum_of_squaresHave a look at the Example code for this chapter
Nothing to many¶
Normally we would use @originate to create files from scratch, for example at the beginning of the pipeline.
However, sometimes, it is not possible to determine ahead of time how many files you will be creating from scratch. @split can also be useful even in such cases:
from random import randint from ruffus import * import os # Create between 2 and 5 files @split(None, "*.start") def create_initial_files(no_input_file, output_files): # cleanup first for oo in output_files: os.unlink(oo) # make new files for ii in range(randint(2,5)): open("%d.start" % ii, "w") @transform(create_initial_files, suffix(".start"), ".processed") def process_files(input_file, output_file): open(output_file, "w") pipeline_run()Giving:
>>> pipeline_run() Job = [None -> *.start] completed Completed Task = create_initial_files Job = [0.start -> 0.processed] completed Job = [1.start -> 1.processed] completed Completed Task = process_files