FAQ¶
Citations¶
Q. How should Ruffus be cited in academic publications?¶
The official publication describing the original version of Ruffus is:
Leo Goodstadt (2010) : Ruffus: a lightweight Python library for computational pipelines. Bioinformatics 26(21): 2778-2779
Good practices¶
Q. What is the best way of keeping my data and workings separate?¶
It is good practice to run your pipeline in a temporary, “working” directory away from your original data.
The first step of your pipeline might be to make softlinks to your original data in your working directory. This is example (relatively paranoid) code to do just this:
def re_symlink (input_file, soft_link_name, logger, logging_mutex): """ Helper function: relinks soft symbolic link if necessary """ # Guard agains soft linking to oneself: Disastrous consequences of deleting the original files!! if input_file == soft_link_name: logger.debug("Warning: No symbolic link made. You are using the original data directory as the working directory.") return # Soft link already exists: delete for relink? if os.path.lexists(soft_link_name): # do not delete or overwrite real (non-soft link) file if not os.path.islink(soft_link_name): raise Exception("%s exists and is not a link" % soft_link_name) try: os.unlink(soft_link_name) except: with logging_mutex: logger.debug("Can't unlink %s" % (soft_link_name)) with logging_mutex: logger.debug("os.symlink(%s, %s)" % (input_file, soft_link_name)) # # symbolic link relative to original directory so that the entire path # can be moved around with breaking everything # os.symlink( os.path.relpath(os.path.abspath(input_file), os.path.abspath(os.path.dirname(soft_link_name))), soft_link_name) # # First task should soft link data to working directory # @jobs_limit(1) @mkdir(options.working_dir) @transform( input_files, formatter(), # move to working directory os.path.join(options.working_dir, "{basename[0]}{ext[0]}"), logger, logging_mutex ) def soft_link_inputs_to_working_directory (input_file, soft_link_name, logger, logging_mutex): """ Make soft link in working directory """ with logging_mutex: logger.info("Linking files %(input_file)s -> %(soft_link_name)s\n" % locals()) re_symlink(input_file, soft_link_name, logger, logging_mutex)
Q. What is the best way of handling data in file pairs (or triplets etc.)¶
In Bioinformatics, DNA data often consists of only the nucleotide sequence at the two ends of larger fragments. The paired_end or mate pair data frequently consists of of file pairs with conveniently related names such as “.R1.fastq” and “.R2.fastq”.
At some point in data pipeline, these file pairs or triplets must find each other and be analysed in the same job.
Provided these file pairs or triplets are named consistently, an easiest way to regroup them is to use the Ruffus @collate decorator. For example:
@collate(original_data_files, # match file name up to the "R1.fastq.gz" formatter("([^/]+)R[12].fastq.gz$"), # Create output parameter supplied to next task "{path[0]}/{1[0]}.sam", logger, logger_mutex) def handle_paired_end(input_files, output_paired_files, logger, logger_mutex): # check that we really have a pair of two files not an orphaned singleton if len(input_files) != 2: raise Exception("One of read pairs %s missing" % (input_files,)) # do stuff hereThis (incomplete, untested) example code shows what this would look like in vivo.
General¶
Q. Ruffus won’t create dependency graphs¶
A. You need to have installed dot from Graphviz to produce pretty flowcharts likes this:
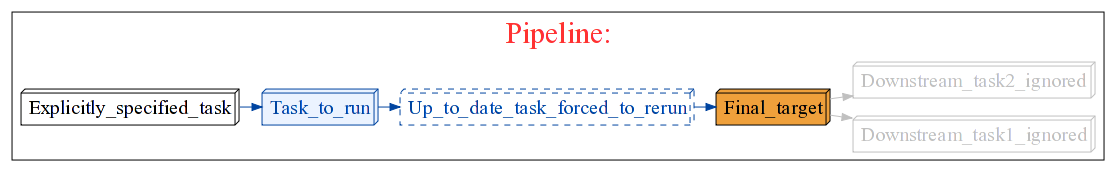
Q. Ruffus seems to be hanging in the same place¶
A. If ruffus is interrupted, for example, by a Ctrl-C, you will often find the following lines of code highlighted:
File "build/bdist.linux-x86_64/egg/ruffus/task.py", line 1904, in pipeline_run File "build/bdist.linux-x86_64/egg/ruffus/task.py", line 1380, in run_all_jobs_in_task File "/xxxx/python2.6/multiprocessing/pool.py", line 507, in next self._cond.wait(timeout) File "/xxxxx/python2.6/threading.py", line 237, in wait waiter.acquire()This is not where ruffus is hanging but the boundary between the main programme process and the sub-processes which run ruffus jobs in parallel.
This is naturally where broken execution threads get washed up onto.
Q. Regular expression substitutions don’t work¶
A. If you are using the special regular expression forms "\1", "\2" etc. to refer to matching groups, remember to ‘escape’ the subsitution pattern string. The best option is to use ‘raw’ python strings. For example:
r"\1_substitutes\2correctly\3four\4times"Ruffus will throw an exception if it sees an unescaped "\1" or "\2" in a file name.
Q. How to force a pipeline to appear up to date?¶
I have made a trivial modification to one of my data files and now Ruffus wants to rerun my month long pipeline. How can I convince Ruffus that everything is fine and to leave things as they are?
The standard way to do what you are trying to do is to touch all the files downstream... That way the modification times of your analysis files would postdate your existing files. You can do this manually but Ruffus also provides direct support:
pipeline_run (touch_files_only = True)pipeline_run will run your script normally stepping over up-to-date tasks and starting with jobs which look out of date. However, after that, none of your pipeline task functions will be called, instead, each non-up-to-date file is touch-ed in turn so that the file modification dates follow on successively.
See the documentation for pipeline_run()
It is even simpler if you are using the new Ruffus.cmdline support from version 2.4. You can just type
your script --touch_files_only [--other_options_of_your_own_etc]See command line documentation.
Q. How can I use my own decorators with Ruffus?¶
(Thanks to Radhouane Aniba for contributing to this answer.)
- With care! If the following two points are observed:
1. Use @wraps from functools or Michele Simionato’s decorator module¶
These will automatically forward attributes from the task function correctly:
- __name__ and __module__ is used to identify functions uniquely in a Ruffus pipeline, and
- pipeline_task is used to hold per task data
2. Always call Ruffus decorators first before your own decorators.¶
Otherwise, your decorator will be ignored.
So this works:
@follows(prev_task) @custom_decorator(something) def test(): passThis is a bit futile
# ignore @custom_decorator @custom_decorator(something) @follows(prev_task) def test(): passThis order dependency is an unfortunate quirk of how python decorators work. The last (rather futile) piece of code is equivalent to:
test = custom_decorator(something)(ruffus.follows(prev_task)(test))Unfortunately, Ruffus has no idea that someone else (custom_decorator) is also modifying the test() function after it (ruffus.follows) has had its go.
Example decorator:¶
Let us look at a decorator to time jobs:
import sys, time def time_func_call(func, stream, *args, **kwargs): """prints elapsed time to standard out, or any other file-like object with a .write() method. """ start = time.time() # Run the decorated function. ret = func(*args, **kwargs) # Stop the timer. end = time.time() elapsed = end - start stream.write("{} took {} seconds\n".format(func.__name__, elapsed)) return ret from ruffus import * import sys import time @time_job(sys.stderr) def first_task(): print "First task" @follows(first_task) @time_job(sys.stderr) def second_task(): print "Second task" @follows(second_task) @time_job(sys.stderr) def final_task(): print "Final task" pipeline_run()What would @time_job look like?
1. Using functools @wraps¶
import functools def time_job(stream=sys.stdout): def actual_time_job(func): @functools.wraps(func) def wrapper(*args, **kwargs): return time_func_call(func, stream, *args, **kwargs) return wrapper return actual_time_job
2. Using Michele Simionato’s decorator module¶
import decorator def time_job(stream=sys.stdout): def time_job(func, *args, **kwargs): return time_func_call(func, stream, *args, **kwargs) return decorator.decorator(time_job)
2. By hand, using a callable object¶
class time_job(object): def __init__(self, stream=sys.stdout): self.stream = stream def __call__(self, func): def inner(*args, **kwargs): return time_func_call(func, self.stream, *args, **kwargs) # remember to forward __name__ inner.__name__ = func.__name__ inner.__module__ = func.__module__ inner.__doc__ = func.__doc__ if hasattr(func, "pipeline_task"): inner.pipeline_task = func.pipeline_task return inner
Q. Can a task function in a Ruffus pipeline be called normally outside of Ruffus?¶
A. Yes. Most python decorators wrap themselves around a function. However, Ruffus leaves the original function untouched and unwrapped. Instead, Ruffus adds a pipeline_task attribute to the task function to signal that this is a pipelined function.
This means the original task function can be called just like any other python function.
Q. My Ruffus tasks create two files at a time. Why is the second one ignored in successive stages of my pipeline?¶
This is my code:
from ruffus import * import sys @transform("start.input", regex(".+"), ("first_output.txt", "second_output.txt")) def task1(i,o): pass @transform(task1, suffix(".txt"), ".result") def task2(i, o): pass pipeline_printout(sys.stdout, [task2], verbose=3)________________________________________ Tasks which will be run: Task = task1 Job = [start.input ->[first_output.txt, second_output.txt]] Task = task2 Job = [[first_output.txt, second_output.txt] ->first_output.result] ________________________________________A: This code produces a single output of a tuple of 2 files. In fact, you want two outputs, each consisting of 1 file.
You want a single job (single input) to be produce multiple outputs (multiple jobs in downstream tasks). This is a one-to-many operation which calls for @split:
from ruffus import * import sys @split("start.input", ("first_output.txt", "second_output.txt")) def task1(i,o): pass @transform(task1, suffix(".txt"), ".result") def task2(i, o): pass pipeline_printout(sys.stdout, [task2], verbose=3)________________________________________ Tasks which will be run: Task = task1 Job = [start.input ->[first_output.txt, second_output.txt]] Task = task2 Job = [first_output.txt ->first_output.result] Job = [second_output.txt ->second_output.result] ________________________________________
Q. How can a Ruffus task produce output which goes off in different directions?¶
A. As above, anytime there is a situation which requires a one-to-many operation, you should reach for @subdivide. The advanced form takes a regular expression, making it easier to produce multiple derivatives of the input file. The following example subdivides 2 jobs each into 3, so that the subsequence task will run 2 x 3 = 6 jobs.
from ruffus import * import sys @subdivide(["1.input_file", "2.input_file"], regex(r"(.+).input_file"), # match file prefix [r"\1.file_type1", r"\1.file_type2", r"\1.file_type3"]) def split_task(input, output): pass @transform(split_task, regex("(.+)"), r"\1.test") def test_split_output(i, o): pass pipeline_printout(sys.stdout, [test_split_output], verbose = 3)Each of the original 2 files have been split in three so that test_split_output will run 6 jobs simultaneously.
________________________________________ Tasks which will be run: Task = split_task Job = [1.input_file ->[1.file_type1, 1.file_type2, 1.file_type3]] Job = [2.input_file ->[2.file_type1, 2.file_type2, 2.file_type3]] Task = test_split_output Job = [1.file_type1 ->1.file_type1.test] Job = [1.file_type2 ->1.file_type2.test] Job = [1.file_type3 ->1.file_type3.test] Job = [2.file_type1 ->2.file_type1.test] Job = [2.file_type2 ->2.file_type2.test] Job = [2.file_type3 ->2.file_type3.test] ________________________________________
Q. Can I call extra code before each job?¶
A. This is easily accomplished by hijacking the process for checking if jobs are up to date or not (@check_if_uptodate):
from ruffus import * import sys def run_this_before_each_job (*args): print "Calling function before each job using these args", args # Remember to delegate to the default *Ruffus* code for checking if # jobs need to run. return needs_update_check_modify_time(*args) @check_if_uptodate(run_this_before_each_job) @files([[None, "a.1"], [None, "b.1"]]) def task_func(input, output): pass pipeline_printout(sys.stdout, [task_func])This results in:
________________________________________ >>> pipeline_run([task_func]) Calling function before each job using these args (None, 'a.1') Calling function before each job using these args (None, 'a.1') Calling function before each job using these args (None, 'b.1') Job = [None -> a.1] completed Job = [None -> b.1] completed Completed Task = task_funcNote
Because run_this_before_each_job(...) is called whenever Ruffus checks to see if a job is up to date or not, the function may be called twice for some jobs (e.g. (None, 'a.1') above).
Q. Does Ruffus allow checkpointing: to distinguish interrupted and completed results?¶
A. Use the builtin sqlite checkpointing¶
By default, pipeline_run(...) will save the timestamps for output files from successfully run jobs to an sqlite database file (.ruffus_history.sqlite) in the current directory .
- If you are using Ruffus.cmdline, you can change the checksum / timestamp database file name on the command line using --checksum_file_name NNNN
The level of timestamping / checksumming can be set via the checksum_level parameter:
pipeline_run(..., checksum_level = N, ...)where the default is 1:
level 0 : Use only file timestamps level 1 : above, plus timestamp of successful job completion level 2 : above, plus a checksum of the pipeline function body level 3 : above, plus a checksum of the pipeline function default arguments and the additional arguments passed in by task decorators
A. Use a flag file¶
When gmake is interrupted, it will delete the target file it is updating so that the target is remade from scratch when make is next run. Ruffus, by design, does not do this because, more often than not, the partial / incomplete file may be usefully if only to reveal, for example, what might have caused an interrupting error or exception. It also seems a bit too clever and underhand to go around the programmer’s back to delete files...
A common Ruffus convention is create an empty checkpoint or “flag” file whose sole purpose is to record a modification-time and the successful completion of a job.
This would be task with a completion flag:
# # Assuming a pipelined task function named "stage1" # @transform(stage1, suffix(".stage1"), [".stage2", ".stage2_finished"] ) def stage2 (input_files, output_files): task_output_file, flag_file = output_files cmd = ("do_something2 %(input_file)s >| %(task_output_file)s ") cmd = cmd % { "input_file": input_files[0], "task_output_file": task_output_file } if not os.system( cmd ): #88888888888888888888888888888888888888888888888888888888888888888888888888888 # # It worked: Create completion flag_file # open(flag_file, "w") # #88888888888888888888888888888888888888888888888888888888888888888888888888888The flag_files xxx.stage2_finished indicate that each job is finished. If this is missing, xxx.stage2 is only a partial, interrupted result.
The only thing to be aware of is that the flag file will appear in the list of inputs of the downstream task, which should accordingly look like this:
@transform(stage2, suffix(".stage2"), [".stage3", ".stage3_finished"] ) def stage3 (input_files, output_files): #888888888888888888888888888888888888888888888888888888888888888888888888888888888 # # Note that the first parameter is a LIST of input files, the last of which # is the flag file from the previous task which we can ignore # input_file, previous_flag_file = input_files # #888888888888888888888888888888888888888888888888888888888888888888888888888888888 task_output_file, flag_file = output_files cmd = ("do_something3 %(input_file)s >| %(task_output_file)s ") cmd = cmd % { "input_file": input_file, "task_output_file": task_output_file } # completion flag file for this task if not os.system( cmd ): open(flag_file, "w")The Bioinformatics example contains code for checkpointing.
A. Use a temp file¶
Thanks to Martin Goodson for suggesting this and providing an example. In his words:
“I normally use a decorator to create a temporary file which is only renamed after the task has completed without any problems. This seems a more elegant solution to the problem:”
def usetemp(task_func): """ Decorate a function to write to a tmp file and then rename it. So half finished tasks cannot create up to date targets. """ @wraps(task_func) def wrapper_function(*args, **kwargs): args=list(args) outnames=args[1] if not isinstance(outnames, basestring) and hasattr(outnames, '__getitem__'): tmpnames=[str(x)+".tmp" for x in outnames] args[1]=tmpnames task_func(*args, **kwargs) try: for tmp, name in zip(tmpnames, outnames): if os.path.exists(tmp): os.rename(tmp, str(name)) except BaseException as e: for name in outnames: if os.path.exists(name): os.remove(name) raise (e) else: tmp=str(outnames)+'.tmp' args[1]=tmp task_func(*args, **kwargs) os.rename(tmp, str(outnames)) return wrapper_functionUse like this:
@files(None, 'client1.price') @usetemp def getusers(inputfile, outputname): #************************************************** # code goes here # outputname now refers to temporary file pass
Windows¶
Q. Windows seems to spawn ruffus processes recursively¶
A. It is necessary to protect the “entry point” of the program under windows. Otherwise, a new process will be started each time the main module is imported by a new Python interpreter as an unintended side effects. Causing a cascade of new processes.
See: http://docs.python.org/library/multiprocessing.html#multiprocessing-programming
This code works:
if __name__ == '__main__': try: pipeline_run([parallel_task], multiprocess = 5) except Exception, e: print e.args
Sun Grid Engine / PBS / SLURM etc¶
Q. Can Ruffus be used to manage a cluster or grid based pipeline?¶
- Some minimum modifications have to be made to your Ruffus script to allow it to submit jobs to a cluster
Thanks to Andreas Heger and others at CGAT and Bernie Pope for contributing ideas and code.
Q. When I submit lots of jobs via Sun Grid Engine (SGE), the head node occassionally freezes and dies¶
- You need to use multithreading rather than multiprocessing. See ruffus.drmaa_wrapper
Q. Keeping Large intermediate files¶
Sometimes pipelines create a large number of intermediate files which might not be needed later.
Unfortunately, the current design of Ruffus requires these files to hang around otherwise the pipeline will not know that it ran successfully.
We have some tentative plans to get around this but in the meantime, Bernie Pope suggests truncating intermediate files in place, preserving timestamps:
# truncate a file to zero bytes, and preserve its original modification time def zeroFile(file): if os.path.exists(file): # save the current time of the file timeInfo = os.stat(file) try: f = open(file,'w') except IOError: pass else: f.truncate(0) f.close() # change the time of the file back to what it was os.utime(file,(timeInfo.st_atime, timeInfo.st_mtime))
Sharing python objects between Ruffus processes running concurrently¶
The design of Ruffus envisages that much of the data flow in pipelines occurs in files but it is also possible to pass python objects in memory.
Ruffus uses the multiprocessing module and much of the following is a summary of what is covered in depth in the Python Standard Library Documentation.
Running Ruffus using pipeline_run(..., multiprocess = NNN) where NNN > 1 runs each job concurrently on up to NNN separate local processes. Each task function runs independently in a different python intepreter, possibly on a different CPU, in the most efficient way. However, this does mean we have to pay some attention to how data is sent across process boundaries (unlike the situation with pipeline_run(..., multithread = NNN) ).
The python code and data which comprises your multitasking Ruffus job is sent to a separate process in three ways:
- The python function code and data objects are pickled, i.e. converting into a byte stream, by the master process, sent to the remote process before being converted back into normal python (unpickling).
- The parameters for your jobs, i.e. what Ruffus calls your task functions with, are separately pickled and sent to the remote process via multiprocessing.Queue
- You can share and synchronise other data yourselves. The canonical example is the logger provided by Ruffus.cmdline.setup_logging
Note
Check that your function code and data can be pickled.
Only functions, built-in functions and classes defined at the top level of a module are picklable.
The following answers are a short “how-to” for sharing and synchronising data yourselves.
Why am I getting PicklingError?¶
What is happening? Didn’t Joan of Arc solve this once and for all?
Some of the data or code in your function cannot be pickled and is being asked to be sent by python mulitprocessing across process boundaries.
When you run your pipeline using multiprocess:
pipeline_run([], verbose = 5, multiprocess = 5, logger = ruffusLoggerProxy)You will get the following errors:
Exception in thread Thread-2: Traceback (most recent call last): File "/path/to/python/python2.7/threading.py", line 808, in __bootstrap_inner self.run() File "/path/to/python/python2.7/threading.py", line 761, in run self.__target(*self.__args, * *self.__kwargs) File "/path/to/python/python2.7/multiprocessing/pool.py", line 342, in _handle_tasks put(task) PicklingError: Can't pickle <type 'function'>: attribute lookup __builtin__.function failedwhich go away when you set pipeline_run([], multiprocess = 1, ...)
Unfortunately, pickling errors are particularly ill-served by standard python error messages. The only really good advice is to take the offending code and try and pickle it yourself and narrow down the errors. Check your objects against the list in the pickle module. Watch out especially for nested functions. These will have to be moved to file scope. Other objects may have to be passed in proxy (see below).
How about synchronising python objects in real time?¶
- You can use managers and proxy objects from the multiprocessing module.
The underlying python object would be owned and managed by a (hidden) server process. Other processes can access the shared objects transparently by using proxies. This is how the logger provided by Ruffus.cmdline.setup_logging works:
# optional logger which can be passed to ruffus tasks logger, logger_mutex = cmdline.setup_logging (__name__, options.log_file, options.verbose)logger is a proxy for the underlying python logger object, and it can be shared freely between processes.
The best course is to pass logger as a parameter to a Ruffus task.
The only caveat is that we should make sure multiple jobs are not writting to the log at the same time. To synchronise logging, we use proxy to a non-reentrant multiprocessing.lock.
logger, logger_mutex = cmdline.setup_logging (__name__, options.log_file, options.verbose) @transform(previous_task, suffix(".foo"), ".bar", logger, logger_mutex) def next_task(input_file, output_file, logger, logger_mutex): with logger_mutex: logger.info("We are in the middle of next_task: %s -> %s" % (input_file, output_file))
How do I send python objects back and forth without tangling myself in horrible synchronisation code?¶
- Sharing python objects by passing messages is a much more modern and safer way to coordinate multitasking than using synchronization primitives like locks.
The python multiprocessing module provides support for passing python objects as messages between processes. You can either use pipes or queues. The idea is that one process pushes and object onto a pipe or queue and the other processes pops it out at the other end. Pipes are only two ended so queues are usually a better fit for sending data to multiple Ruffus jobs.
Proxies for queues can be passed between processes as in the previous section