Chapter 8: Specifying output file names with formatter() and regex()¶
See also
- Manual Table of Contents
- suffix() syntax
- formatter() syntax
- regex() syntax
Note
Remember to look at the example code:
Review¶
Computational pipelines transform your data in stages until the final result is produced. The most straightforward way to use Ruffus is to hold the intermediate results after each stage in a series of files with related file names.
Part of telling Ruffus how these pipeline stages or task functions are connected together is to write simple rules for how to the file names for each stage follow on from each other. Ruffus helps you to specify these file naming rules.
Note
The best way to design a pipeline is to:
- Write down the file names of the data as it flows across your pipeline. Do these file names follow a pattern ?
- Write down the names of functions which transforms the data at each stage of the pipeline.
A different file name suffix() for each pipeline stage¶
The easiest and cleanest way to write Ruffus pipelines is to use a different suffix for each stage of your pipeline.
We used this approach in Chapter 1: An introduction to basic Ruffus syntax and in code from Chapter 3: More on @transform-ing data:
#Task Name: File suffices _________________________ ______________________ create_initial_file_pairs *.start first_task *.output.1 second_task *.output.2There is a long standing convention of using file suffices to denote file type: For example, a “compile” task might convert source files of type *.c to object files of type *.o.
- We can think of Ruffus tasks comprising :
- recipes in @transform(...) for transforming file names: changing .c to a .o (e.g. AA.c -> AA.o BB.c -> BB.o)
- recipes in a task function def foo_bar() for transforming your data: from source .c to object .o
Let us review the Ruffus syntax for doing this:
@transform( create_initial_file_pairs, # Input: Name of previous task(s) suffix(".start"), # Matching suffix ".output.1") # Replacement string def first_task(input_files, output_file): with open(output_file, "w"): pass
Input:
- The first parameter for @transform can be a mixture of one or more:
- previous tasks (e.g. create_initial_file_pairs)
- file names (all python strings are treated as paths)
- glob specifications (e.g *.c, /my/path/*.foo)
Each element provides an input for the task. So if the previous task create_initial_file_pairs has five outputs, the next @transform task will accept these as five separate inputs leading to five independent jobs.
The second parameter suffix(".start") must match the end of the first string in each input. For example, create_initial_file_pairs produces the list ['job1.a.start', 'job1.b.start'], then suffix(".start") must matches the first string, i.e. 'job1.a.start'. If the input is nested structure, this would be iterated through recursively to find the first string.
Note
Inputs which do not match the suffix are discarded altogether.
Replacement:
The third parameter is the replacement for the suffix. The pair of input strings in the step3 example produces the following output parameter
input_parameters = ['job1.a.start', 'job1.b.start'] matching_input = 'job1.a.start' output_parameter = 'job1.a.output.1'When the pipeline is run, this results in the following equivalent call to first_task(...):
first_task(['job1.a.start', 'job1.b.start'], 'job1.a.output.1'):The replacement parameter can itself be a list or any arbitrary complicated structure:
@transform(create_initial_file_pairs, # Input suffix(".a.start"), # Matching suffix [".output.a.1", ".output.b.1", 45]) # Replacement list def first_task(input_files, output_parameters): print "input_parameters = ", input_files print "output_parameters = ", output_parametersIn which case, all the strings are used as replacements, other values are left untouched, and we obtain the following:
# job #1 input = ['job1.a.start', 'job1.b.start'] output = ['job1.output.a.1', 'job1.output.b.1', 45] # job #2 input = ['job2.a.start', 'job2.b.start'] output = ['job2.output.a.1', 'job2.output.b.1', 45] # job #3 input = ['job3.a.start', 'job3.b.start'] output = ['job3.output.a.1', 'job3.output.b.1', 45]Note how task function is called with the value 45 verbatim because it is not a string.
formatter() manipulates pathnames and regular expression¶
suffix() replacement is the cleanest and easiest way to generate suitable output file names for each stage in a pipeline. Often, however, we require more complicated manipulations to specify our file names. For example,
- It is common to have to change directories from a data directory to a working directory as the first step of a pipeline.
- Data management can be simplified by separate files from each pipeline stage into their own directory.
- Information may have to be decoded from data file names, e.g. "experiment373.IBM.03March2002.txt"
Though formatter() is much more powerful, the principle and syntax are the same: we take string elements from the Input and perform some replacements to generate the Output parameters.
- Allows easy manipulation of path subcomponents in the style of os.path.split(), and os.path.basename
- Uses familiar python string.format syntax (See string.format examples. )
- Supports optional regular expression (re) matches including named captures.
- Can refer to any file path (i.e. python string) in each input and is not limited like suffix() to the first string.
- Can even refer to individual letters within a match
Path name components¶
formatter() breaks down each input pathname into path name components which can then be recombined in whichever way by the replacement string.
Given an example string of :
input_string = "/directory/to/a/file.name.ext" formatter()the path components are:
- basename: The base name excluding extension, "file.name"
- ext : The extension, ".ext"
- path : The dirname, "/directory/to/a"
- subdir : A list of sub-directories in the path in reverse order, ["a", "to", "directory", "/"]
- subpath : A list of descending sub-paths in reverse order, ["/directory/to/a", "/directory/to", "/directory", "/"]
The replacement string refers to these components by using python string.format style curly braces. "{NAME}"
We refer to an element from the Nth input string by index, for example:
- "{ext[0]}" is the extension of the first file name string in Input.
- "{basename[1]}" is the basename of the second file name in Input.
- "{basename[1][0:3]}" are the first three letters from the basename of the second file name in Input.
subdir, subpath were designed to help you navigate directory hierachies with the minimum of fuss. For example, you might want to graft a hierachical path to another location: "{subpath[0][2]}/from/{subdir[0][0]}/{basename[0]}" neatly replaces just one directory ("to") in the path with another ("from"):
replacement_string = "{subpath[0][2]}/from/{subdir[0][0]}/{basename[0]}" input_string = "/directory/to/a/file.name.ext" result_string = "/directory/from/a/file.name.ext"
Filter and parse using regular expressions¶
Regular expression matches can be used with the similar syntax. Our example string can be parsed using the following regular expression:
input_string = "/directory/to/a/file.name.ext" formatter(r"/directory/(.+)/(?P<MYFILENAME>)\.ext")We capture part of the path using (.+), and the base name using (?P<MYFILENAME>). These matching subgroups can be referred to by index but for greater clarity the second named capture can also be referred to by name, i.e. {MYFILENAME}.
The regular expression components for the first string can thus be referred to as follows:
- {0[0]} : The entire match captured by index, "/directory/to/a/file.name.ext"
- {1[0]} : The first match captured by index, "to/a"
- {2[0]} : The second match captured by index, "file.name"
- {MYFILENAME[0]} : The match captured by name, "file.name"
If each input consists of a list of paths such as ['job1.a.start', 'job1.b.start', 'job1.c.start'], we can match each of them separately by using as many regular expressions as necessary. For example:
input_string = ['job1.a.start', 'job1.b.start', 'job1.c.start'] # Regular expression matches for 1st, 2nd but not 3rd element formatter(".+a.start", "b.start$")Or if you only wanted regular expression matches for the second file name (string), pad with None:
input_string = ['job1.a.start', 'job1.b.start', 'job1.c.start'] # Regular expression matches for 2nd but not 1st or 3rd elements formatter(None, "b.start$")
Using @transform() with formatter()¶
We can put these together in the following example:
from ruffus import * # create initial files @originate([ ['job1.a.start', 'job1.b.start'], ['job2.a.start', 'job2.b.start'], ['job3.a.start', 'job3.c.start'] ]) def create_initial_file_pairs(output_files): # create both files as necessary for output_file in output_files: with open(output_file, "w") as oo: pass #--------------------------------------------------------------- # # formatter # # first task @transform(create_initial_file_pairs, # Input formatter(".+/job(?P<JOBNUMBER>\d+).a.start", # Extract job number ".+/job[123].b.start"), # Match only "b" files ["{path[0]}/jobs{JOBNUMBER[0]}.output.a.1", # Replacement list "{path[1]}/jobs{JOBNUMBER[0]}.output.b.1", 45]) def first_task(input_files, output_parameters): print "input_parameters = ", input_files print "output_parameters = ", output_parameters # # Run # pipeline_run(verbose=0)This produces:
input_parameters = ['job1.a.start', 'job1.b.start'] output_parameters = ['/home/lg/src/temp/jobs1.output.a.1', '/home/lg/src/temp/jobs1.output.b.1', 45] input_parameters = ['job2.a.start', 'job2.b.start'] output_parameters = ['/home/lg/src/temp/jobs2.output.a.1', '/home/lg/src/temp/jobs2.output.b.1', 45]Notice that job3 has 'job3.c.start' as the second file. This fails to match the regular expression and is discarded.
Note
Failed regular expression mismatches are ignored.
formatter() regular expressions are thus very useful in filtering out all files which do not match your specified criteria.
If your some of your task inputs have a mixture of different file types, a simple Formatter(".txt$"), for example, will make your code a lot simpler...
string substitution for “extra” arguments¶
The first two arguments for Ruffus task functions are special because they are the Input and Output parameters which link different stages of a pipeline.
Python strings in these arguments are names of data files whose modification times indicate whether the pipeline is up to date or not.
Other arguments to task functions are not passed down the pipeline but consumed. Any python strings they contain do not need to be file names. These extra arguments are very useful for passing data to pipelined tasks, such as shared values, loggers, programme options etc.
One helpful feature is that strings in these extra arguments are also subject to formatter() string substitution. This means you can leverage the parsing capabilities of Ruffus to decode any information about the pipeline data files, These might include the directories you are running in and parts of the file name.
For example, if we would want to know which files go with which “job number” in the previous example:
from ruffus import * # create initial files @originate([ ['job1.a.start', 'job1.b.start'], ['job2.a.start', 'job2.b.start'], ['job3.a.start', 'job3.c.start'] ]) def create_initial_file_pairs(output_files): for output_file in output_files: with open(output_file, "w") as oo: pass #--------------------------------------------------------------- # # print job number as an extra argument # # first task @transform(create_initial_file_pairs, # Input formatter(".+/job(?P<JOBNUMBER>\d+).a.start", # Extract job number ".+/job[123].b.start"), # Match only "b" files ["{path[0]}/jobs{JOBNUMBER[0]}.output.a.1", # Replacement list "{path[1]}/jobs{JOBNUMBER[0]}.output.b.1"], "{JOBNUMBER[0]}" def first_task(input_files, output_parameters, job_number): print job_number, ":", input_files pipeline_run(verbose=0)>>> pipeline_run(verbose=0) 1 : ['job1.a.start', 'job1.b.start'] 2 : ['job2.a.start', 'job2.b.start']
Changing directories using formatter() in a zoo...¶
Here is a more fun example. We would like to feed the denizens of a zoo. Unfortunately, the file names for these are spread over several directories. Ideally, we would like their food supply to be grouped more sensibly. And, of course, we only want to feed the animals, not the plants.
I have colour coded the input and output files for this task to show how we would like to rearrange them:
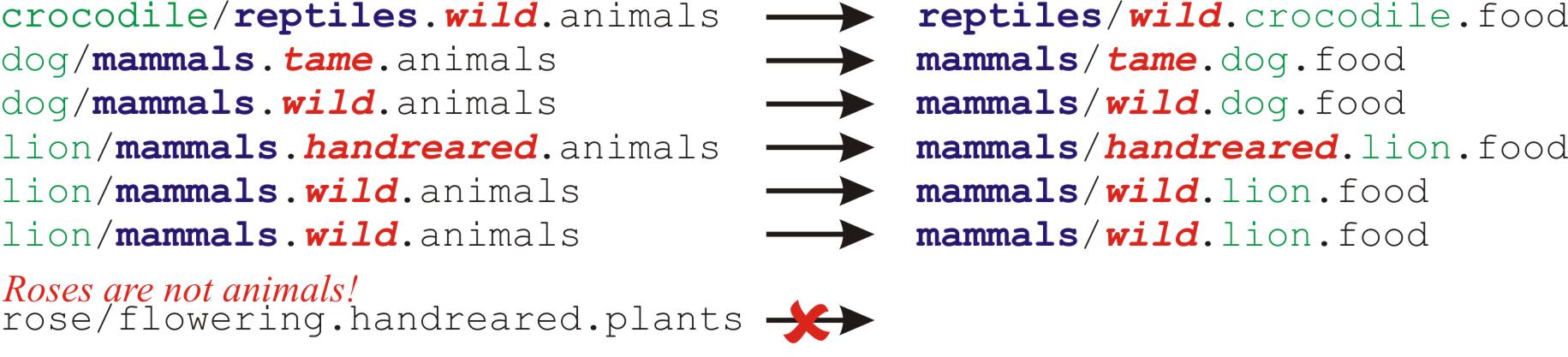 from ruffus import * # Make directories @mkdir(["tiger", "lion", "dog", "crocodile", "rose"]) @originate( # List of animals and plants [ "tiger/mammals.wild.animals", "lion/mammals.wild.animals", "lion/mammals.handreared.animals", "dog/mammals.tame.animals", "dog/mammals.wild.animals", "crocodile/reptiles.wild.animals", "rose/flowering.handreared.plants"]) def create_initial_files(output_file): with open(output_file, "w") as oo: pass # Put different animals in different directories depending on their clade @transform(create_initial_files, # Input formatter(".+/(?P<clade>\w+).(?P<tame>\w+).animals"), # Only animals: ignore plants! "{subpath[0][1]}/{clade[0]}/{tame[0]}.{subdir[0][0]}.food", # Replacement "{subpath[0][1]}/{clade[0]}", # new_directory "{subdir[0][0]}", # animal_name "{tame[0]}") # tameness def feed(input_file, output_file, new_directory, animal_name, tameness): print "Food for the {tameness:11s} {animal_name:9s} = {output_file:90s} will be placed in {new_directory}".format(**locals()) pipeline_run(verbose=0)
from ruffus import * # Make directories @mkdir(["tiger", "lion", "dog", "crocodile", "rose"]) @originate( # List of animals and plants [ "tiger/mammals.wild.animals", "lion/mammals.wild.animals", "lion/mammals.handreared.animals", "dog/mammals.tame.animals", "dog/mammals.wild.animals", "crocodile/reptiles.wild.animals", "rose/flowering.handreared.plants"]) def create_initial_files(output_file): with open(output_file, "w") as oo: pass # Put different animals in different directories depending on their clade @transform(create_initial_files, # Input formatter(".+/(?P<clade>\w+).(?P<tame>\w+).animals"), # Only animals: ignore plants! "{subpath[0][1]}/{clade[0]}/{tame[0]}.{subdir[0][0]}.food", # Replacement "{subpath[0][1]}/{clade[0]}", # new_directory "{subdir[0][0]}", # animal_name "{tame[0]}") # tameness def feed(input_file, output_file, new_directory, animal_name, tameness): print "Food for the {tameness:11s} {animal_name:9s} = {output_file:90s} will be placed in {new_directory}".format(**locals()) pipeline_run(verbose=0)We can see that the food for each animal are now grouped by clade in the same directory, which makes a lot more sense...
Note how we used subpath[0][1] to move down one level of the file path to build a new file name.
>>> pipeline_run(verbose=0) Food for the wild crocodile = ./reptiles/wild.crocodile.food will be placed in ./reptiles Food for the tame dog = ./mammals/tame.dog.food will be placed in ./mammals Food for the wild dog = ./mammals/wild.dog.food will be placed in ./mammals Food for the handreared lion = ./mammals/handreared.lion.food will be placed in ./mammals Food for the wild lion = ./mammals/wild.lion.food will be placed in ./mammals Food for the wild tiger = ./mammals/wild.tiger.food will be placed in ./mammals
regex() manipulates via regular expressions¶
If you are a hard core regular expressions fan, you may want to use regex() instead of suffix() or formatter().
Note
regex() uses regular expressions like formatter() but
- It only matches the first file name in the input. As described above, formatter() can match any one or more of the input filename strings.
- It does not understand file paths so you may have to perform your own directory / file name parsing.
- String replacement uses syntax borrowed from re.sub(), rather than building a result from parsed regular expression (and file path) components
In general formatter() is more powerful and was introduced from version 2.4 is intended to be a more user friendly replacement for regex().
Let us see how the previous zoo example looks with regex():
formatter() code:
# Put different animals in different directories depending on their clade @transform(create_initial_files, # Input formatter(".+/(?P<clade>\w+).(?P<tame>\w+).animals"), # Only animals: ignore plants! "{subpath[0][1]}/{clade[0]}/{tame[0]}.{subdir[0][0]}.food", # Replacement "{subpath[0][1]}/{clade[0]}", # new_directory "{subdir[0][0]}", # animal_name "{tame[0]}") # tameness def feed(input_file, output_file, new_directory, animal_name, tameness): print "Food for the {tameness:11s} {animal_name:9s} = {output_file:90s} will be placed in {new_directory}".format(**locals())regex() code:
# Put different animals in different directories depending on their clade @transform(create_initial_files, # Input regex(r"(.*?/?)(\w+)/(?P<clade>\w+).(?P<tame>\w+).animals"), # Only animals: ignore plants! r"\1/\g<clade>/\g<tame>.\2.food", # Replacement r"\1/\g<clade>", # new_directory r"\2", # animal_name "\g<tame>") # tameness def feed(input_file, output_file, new_directory, animal_name, tameness): print "Food for the {tameness:11s} {animal_name:9s} = {output_file:90s} will be placed in {new_directory}".format(**locals())The regular expression to parse the input file path safely was a bit hairy to write, and it is not clear that it handles all edge conditions (e.g. files in the root directory). Apart from that, if the limitations of regex() do not preclude its use, then the two approaches are not so different in practice.