Chapter 2: Transforming data in a pipeline with @transform¶
See also
Note
Remember to look at the example code:
Review¶
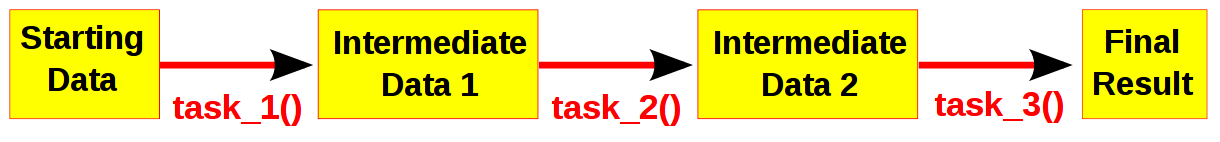
Computational pipelines transform your data in stages until the final result is produced. Ruffus automates the plumbing in your pipeline. You supply the python functions which perform the data transformation, and tell Ruffus how these pipeline stages or task functions are connected together.
Note
The best way to design a pipeline is to:
- write down the file names of the data as it flows across your pipeline
- write down the names of functions which transforms the data at each stage of the pipeline.
Task functions as recipes¶
Each task function of the pipeline is a recipe or rule which can be applied repeatedly to our data.
For example, one can have
- a compile() task which will compile any number of source code files, or
- a count_lines() task which will count the number of lines in any file or
- an align_dna() task which will align the DNA of many chromosomes.
@transform is a 1 to 1 operation¶
@transform is a 1:1 operation because for each input, it generates one output.
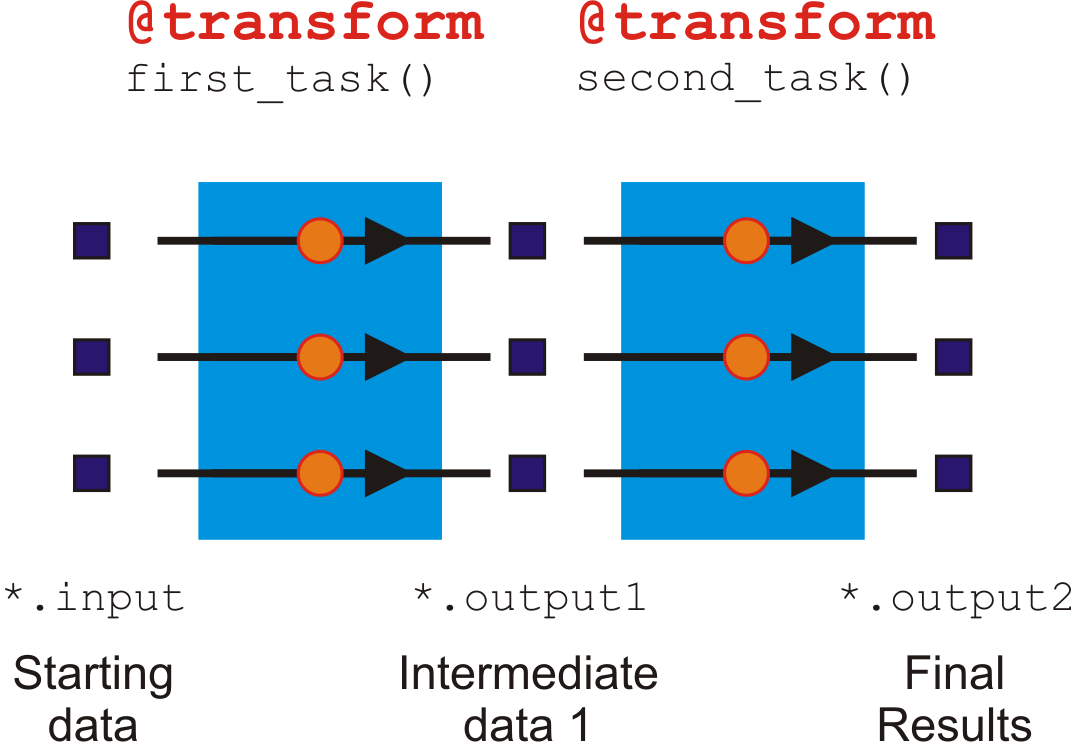
This is obvious when you count the number of jobs at each step. In our example pipeline, there are always three jobs moving through in step at each stage (task).
Each Input or Output is not limited, however, to a single filename. Each job can accept, for example, a pair of files as its Input, or generate more than one file or a dictionary or numbers as its Output.
When each job outputs a pair of files, this does not generate two jobs downstream. It just means that the successive task in the pipeline will receive a list or tuple of files as its input parameter.
Note
The different sort of decorators in Ruffus determine the topology of your pipeline, i.e. how the jobs from different tasks are linked together seamlessly.
@transform always generates one Output for one Input.
In the later parts of the tutorial, we will encounter more decorators which can split up, or join together or group inputs.
In other words, using other decorators Input and Output can have many to one, many to many etc. relationships.
A pair of files as the Input¶
Let us rewrite our previous example so that the Input of the first task are matching pairs of DNA sequence files, processed in tandem.
from ruffus import * starting_files = [("a.1.fastq", "a.2.fastq"), ("a.1.fastq", "a.2.fastq"), ("a.1.fastq", "a.2.fastq")] # # STAGE 1 fasta->sam # @transform(starting_files, # Input = starting files suffix(".1.fastq"), # suffix = .1.fastq ".sam") # Output suffix = .sam def map_dna_sequence(input_files, output_file): # remember there are two input files now ii1 = open(input_files[0]) ii2 = open(input_files[1]) oo = open(output_file, "w")The only changes are to the first task:
pipeline_run() Job = [[a.1.fastq, a.2.fastq] -> a.sam] completed Job = [[a.1.fastq, a.2.fastq] -> a.sam] completed Job = [[a.1.fastq, a.2.fastq] -> a.sam] completed Completed Task = map_dna_sequencesuffix always matches only the first file name in each Input.
Input and Output parameters¶
Ruffus chains together different tasks by taking the Output from one job and plugging it automatically as the Input of the next.
The first two parameters of each job are the Input and Output parameters respectively.
In the above example, we have:
>>> pipeline_run() Job = [a.bam -> a.statistics, use_linear_model] completed Job = [b.bam -> b.statistics, use_linear_model] completed Job = [c.bam -> c.statistics, use_linear_model] completed Completed Task = summarise_bam_file
Parameters for summarise_bam_file() Inputs Outputs Extra "a.bam" "a.statistics" "use_linear_model" "b.bam" "b.statistics" "use_linear_model" "c.bam" "c.statistics" "use_linear_model" Extra parameters are for the consumption of summarise_bam_file() and will not passed to the next task.
Ruffus was designed for pipelines which save intermediate data in files. This is not compulsory but saving your data in files at each step provides many advantages:
- Ruffus can use file system time stamps to check if your pipeline is up to date
- Your data is persistent across runs
- This is a good way to pass large amounts of data across processes and computational nodes
Nevertheless, all the task parameters can include anything which suits your workflow, from lists of files, to numbers, sets or tuples. Ruffus imposes few constraints on what you would like to send to each stage of your pipeline.
Ruffus does, however, assume that if the Input and Output parameter contains strings, these will be interpreted as file names required by and produced by that job. As we shall see, the modification times of these file names indicate whether that part of the pipeline is up to date or needs to be rerun.